
Stuck at home: the puzzle of a locally-abundant, thermophilic bacterial genus that has not dispersed beyond Aotearoa-New Zealand
2023-2026
Role: Principal Investigator
Funder: Marsden Fund (RSNZ)
Students: Holly Welford, Meghan Marshall, Natasha Korvin
What makes Venenivibrio a microbial “manu”?
Geographic isolation enables the evolution of new organisms. Some species develop traits that allow them to migrate and populate widespread regions of the globe, while others are limited (endemic) to localised areas.
The bacterial genus Venenivibrio is the most abundant microorganism across Aotearoa-New Zealand hot springs, but it isn’t known anywhere else in the world. In contrast, its sister genus Sulfurihydrogenibium, which has the same broad physiological characteristics, is readily identified in hot springs outside Aotearoa-New Zealand.
This project is investigating how Venenivibrio distributes domestically and why it cannot, or does not, survive long distance migration. In collaboration with Ngāti Tahu-Ngāti Whaoa (mana whenua of the type strain), we hope to learn what makes this species a taonga. The moniker “microbial manu” (bird) was suggested by Evelyn Forrest (Ngāti Tahu-Ngāti Whaoa Rūnanga Trust).
Publications linked to this project: [1, 2, and 3].

In extremis – revealing novel metabolic pathways that support microbial populations in the deep subsurface biosphere of Mt Erebus, Antarctica
2019-2025
Role: Co-Principal Investigator (co-PI: Prof. Craig Cary – UoW)
Funder: Marsden Fund (RSNZ)
What can the subsurface geothermal habitats on Mt. Erebus (Antarctica) teach us about the ecology of the global deep subsurface biosphere?
Investigating microorganisms adapted to geothermal environments has been critically important to understanding the physical and chemical constraints on life. The geothermal system on Mt. Erebus, Antarctica, is geographically isolated and geochemically unique. We are proposing that the subsurface rock matrix of this geothermal system naturally enriches for novel microbial communities with rare/undescribed metabolisms.
This project explores novel catabolic pathways to reveal physicochemical, genomic, and bioenergetic knowledge that is unknown yet theoretically feasible. Our findings will potentially re-define the bioenergetic boundaries of life on Earth and address questions on the evolution of microbial energetic pathways and diversity of life they support.
Publications linked to this project: [1], [2] and [3]

From geothermal hot springs to microbial gene pools: Explaining intra-species genomic variations in bacteria
2018-2021
Role: Associate Investigator (PI: Dr Charlie Lee – UoW)
Funder: Marsden Fund (RSNZ)
Leveraging New Zealand’s unique geothermal microbial communities, we used comparative genomics to explore a new concept of microbial species.
In the conventional framework for evolution, natural selection acts on individual organisms exhibiting traits rather than directly on trait-conferring genes. But natural bacterial populations exhibit intra-species genomic variations. Genetically and ecologically cohesive populations within natural microbial communities cannot be reliably and objectively identified and tracked.
This project explicitly considered horizontal gene transfer, which is prevalent in bacteria, when defining bacterial “species”. The ecology of geothermal springs was used as a model system to examine how ecological populations of bacteria and archaea might allow microbial “species” to be delineated exclusively through discontinuities in gene flow.
Publications linked to this project: Coming Soon.

The 1000 Springs Project: A microbial bioinventory of geothermal ecosystems
2012-2015
Role: Co-Principal Investigator (co-PI: Prof. Craig Cary, UoW)
Funder: MBIE Smart Idea
Students: Jean Power, Caitlin Lowe
A bioinventory of the microbial diversity of Aotearoa’s geothermal ecosystems.
Geothermal ecosystems are globally rare and an iconic feature of the Aoteaora-New Zealand landscape. Yet, little is known about the unique populations of microorganisms (Bacteria and Archaea) which inhabit these environments or the ecological conditions that support them.
We sampled a broad cross section of physicochemical conditions presented by geothermally-influenced springs within the Taupō Volcanic Zone. Over 1000 samples were collected form the water columns of the springs, most from individual features, as well as some repeated sampling for a longer term temporal study or for quality control reasons. The 1000 Springs website catalogues the microbial biodiversity and geochemical information from this research
Publications linked to this project: [1], [2], [3], [4] and [5]
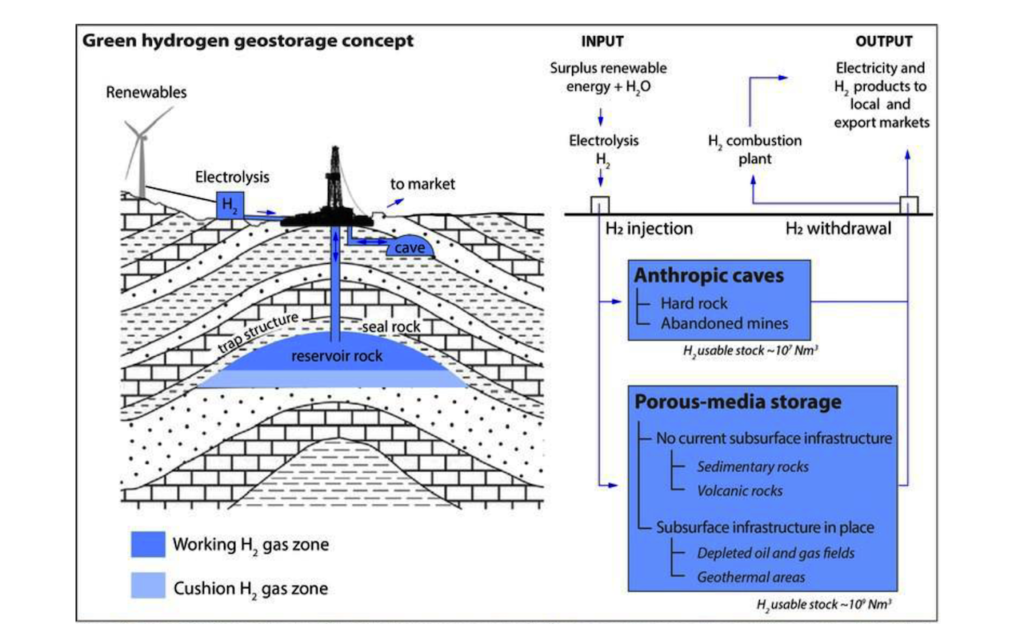
Pūhiko Nukutū: a green hydrogen geostorage battery in Taranaki
2022-2027
Role: Associate Investigator (PI: Prof. Andy Nicol, UoC)
Funder: MBIE Research Programme
Students: Kelsey McKenzie
Pūhiko Nukutū (Earth Battery) is investigating how to create large stores of green hydrogen underground earth batteries.
In this project, our focus is on the deep biosphere microbial communities. We are examining the complex interactions of rocks and microbes when exposed to hydrogen (H2), to predict how, where and for how long hydrogen can be stored.
We are determining the hydrogenotrophic microbial community composition and activity in deep subsurface samples using cultivation and molecular techniques. We will use microcosms to cultivate known hydrogenotrophic strains (methanogens, others) and hydrogen-utilising consortia, in order to determine methane (CH4) generation and H2 consumption rates. Biogeochemical reactions between rock substrates, microorganisms and H2, will be determined using physicochemical analyses of target rock microcosms.
Publications linked to this project: Coming Soon.

Flipping the paradigm: feeding methane to cows
2017-2022
Role: Associate Investigator (PI: Dr Carlo Carere, UoC)
Funder: MBIE Smart Idea
Student: Carlos Cartin Caballero
A biotechnology platform that takes industrial waste gases from geothermal power stations and converts them into protein-rich biomass.
Methane and carbon dioxide are potent greenhouse gases and common by-products from industrial processes. However, when these and other gases are produced, they are generally at concentrations too dilute to be economically useful and are commonly disposed of either by flaring or venting into the atmosphere.
This research utilises the unique characteristics of naturally-occurring indigenous extremophilic microorganisms to capture CH4 and CO2 emissions and transform them into a valuable product. The protein-rich biomass could be used to feed dairy, stock and other farmed animals (including in aquaculture).
Publications linked to this project: [1], [2], [3] and [4]

Te Nohonga Kaitiaki
2018-2020
Role: Associate Investigator (PI: Assoc. Prof. Maui Hudson; Whakatōhea, Ngāruahine and Te Māhaurehure, UoW)
Funder: Genomics Aotearoa
Culturally-informed ethical guidelines for researchers generating genetic information on taonga species or indigenous New Zealand biota.
Genomics Aotearoa aims to build capacity to compile large and complex genomic datasets for the purposes of conservation management and improved breeding in the primary production sector. This requires enhanced research practices to recognise potential Māori rights and interests in taonga species.
This project created guidelines for genomic research with taonga species, including pathways for benefit sharing and commercialisation that explicitly incorporate vision Mātauranga. Engagement process were developed, based on hui experiences and beyond, for Māori communities and genomic research.
Publications linked to this project: [1] and [2]

A ligase-based solution for non-natural nucleic acid synthesis
2022-2025
Role: Associate Investigator (PI: Dr Adele Williamson – UoW)
Funder: MBIE Smart Idea
This project aims to discover and engineer enzymes to build large XNAs from small synthetic pieces.
Xeno-Nucleic-Acids (XNAs) are artificial equivalents of natural genetic material DNA and RNA, and behave in a similar way — folding into double-helices and storing information. But XNAs can have much greater chemical diversity and are often more stable in biological fluids like blood and saliva. This makes XNAs extremely useful for biotechnological applications such as synthetic biology, nanotechnology therapeutics and diagnostics.
To synthesises XNAs, we are using DNA ligases, enzymes that join breaks in double-stranded DNA in nature. We will begin with ligases that we find in the genomes of bacteria and viruses from extreme environments like Antarctica and geothermal regions of Aotearoa-New Zealand.
Publications linked to this project: Coming Soon.
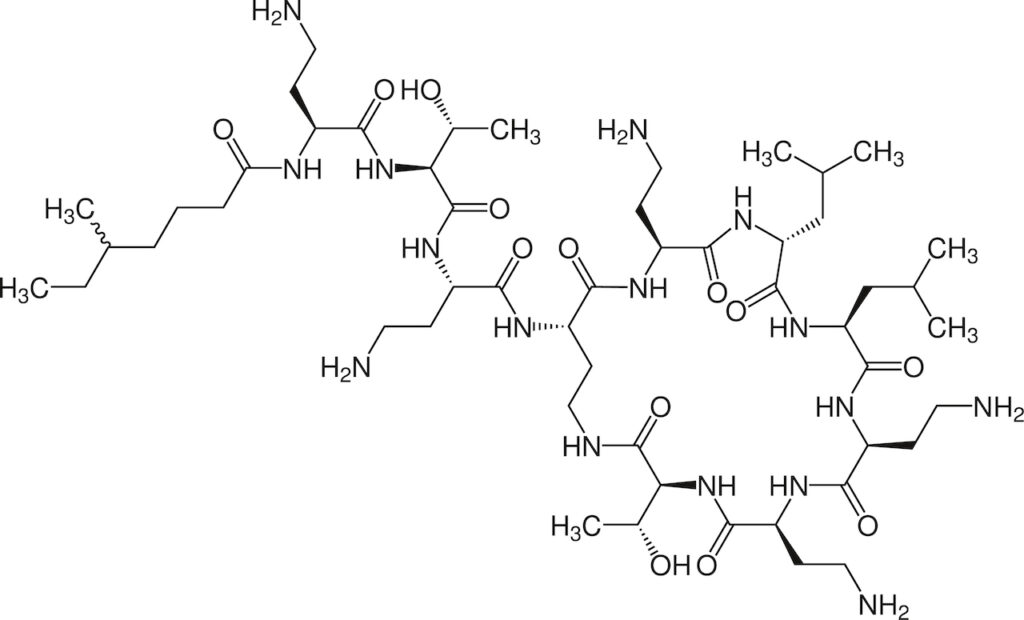
Waerau waikawa iti rongoā paturopi: New Generation Peptide Antibiotics
2020-2025
Role: Associate Investigator (PI: Dame Prof. Margaret Brimble – UoA)
Funder: MBIE Research Programme
Students: Hazel Clemens
This research programme addresses antimicrobial resistance by focusing on knowledge creation for new antibiotics for human use.
Our last lines of chemical defence against multi-drug resistant Gram-negative and Gram-positive bacteria are antimicrobial peptides produced by environmental microbes. Due to their unique bactericidal mechanisms of action, antimicrobial peptides have a lower tendency to elicit antimicrobial resistance than conventional antibiotics.
This programmes develops a transformational drug discovery platform that combines the manufacture of novel lipopeptide antimicrobials, with an innovative NZ microbial genome-mining approach. The genomes of both cultivated and uncultivated New Zealand microbiomes will be explored to discover new lipopeptide scaffolds that possess novel mechanisms of antimicrobial action.
Publications linked to this project: Coming Soon